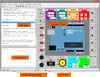
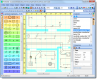
TinkerCell is a Computer-Aided Design software tool for Synthetic Biology. It combines visual interface with programming API (Python, Octave, C, Ruby) and allows users to share their code with each other via a central repository.
TinkerCell is a Computer-Aided Design (CAD) tool for synthetic biology. Since synthetic biology is a rapidly evolving field, here are the issues which were taken into consideration when designing TinkerCell:
- modeling techniques in biology are still at a developmental stage
- experimental techniques are constantly evolving, especially automation and directed evolution methods
- databases of biological components are still maturing
In summary, TinkerCell is designed with the anticipation that the future of synthetic biology will be an intricate interplay between a variety of experimental techniques, databases that store results from experiments, and mathematical models explaining different aspects of the experiments.
The use-case of the ideal CAD program would be like this:
- step 1: user draws a biological system
- step 2: user performs some analysis
- step 3: go back to step 1 if analysis is not satisfactory
The analysis step can potentially include:
- mathematical analysis of non-linear systems
- stochastic simulations, structural analysis, and other methods from systems biology
- prediction of evolutionary trajectories for directed evolution
- analysis and optimization of the DNA sequence
- database look-up to find suitable components
- generate different versions of the same design
- load experimental data from database(s) and relate them to the diagram
- ...more...
It is too much to have all of these functions in TinkerCell from the start. So TinkerCell uses a flexible plug-in framework, which will allow others to contribute new functions to TinkerCell. The plug-ins can be written in C, C++, Python, or Octave. Other languages, e.g. Java, might also get added to this list.
The basic idea behind TinkerCell is to represent a diagram that is detailed enough so that it can be mapped to models or experimental results. A human researcher is able to connect models and experiments because they are able to link them using a conceptual understanding of the system. TinkerCell enables that capability in software.
It is not entirely correct to provide a single mathematical model for a biological system because one mathematical model is just one hypothesis. There can be multiple hypotheses for the same system. TinkerCell distinguishes a “model” from a “diagram”.
While TinkerCell diagrams are very abstract, they also contain all the details needed for different types of analysis. A diagram is not a model itself, but it has enough information for generating one or more models. For example, here is an abstract diagram.
TinkerCell is modular, meaning that each process in the above diagram can consist of subprocesses. In the example diagram above, the enzyme catalysis process has processes within it. The inner-processes can be changed without changing the outer diagram. The illustration below shows how the subprocesses are mapped to the main diagram by identifying the “types” of each component.
Long term goals
The long term goal of TinkerCell is to become one of the key applications involved in efficient engineering of biological (cellular) systems. The way this will work is by connecting TinkerCell to lab automation tools. In this situation, users would draw a system in TinkerCell. Some TinkerCell plugin would send the information to a remote robot. When the results from the experiments return, another TinkerCell plugin would automatically recommend the models to use based on the experimental evidence. In order to enable this pipeline, it is important to represent the initial model is a generic yet detailed format; existing software tools cannot do this, which is why one of the many reasons why pipe is not possible at present.
Current analysis capabilities
- deterministic simulation (using COPASI)
- stochastic simulation (using COPASI)
- steady state computation (using COPASI)
- jacobian computation (using COPASI)
- eigenvalue computation (using COPASI)
- exact stochastic simulation (using COPASI)
- tau-leap based stochastic simulation (using COPASI)
- hybrid simulation (using COPASI)
- stoichiometry matrix structural analysis (using libStructural)
- analysis of model assumptions, such as equilibrium assumptions
- parameter scan (Monte Carlo and steady state analysis)