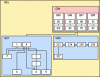
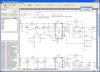
JBioFramework (JBF) is a set of two different chemical separations simulations (2D Electrophoresis and Mass Spectrometry) that are frequently used in chemistry, biochemistry and proteomics research. It is written in the Java programming language and will run on any and all systems that have the JVM installed.
As we continue to develop the software over the coming months/years and attempt to quantify the success of our efforts with testing and reviews, user input is very important. Please don't hesitate to review the software below or email Paul Craig [pac8612@rit.edu] with larger descriptions/bugs/feature ideas.
Our next scheduled release will contain (in addition to 2DE and MassSpec) a 1D Electrophoresis simulation as well as a tab containing ChemAxon's MarvinSketch [http://www.chemaxon.com/products/marvin/] along with some improved functionality. It should be out shortly as we complete the creation and modification of a permanent domain for the project.
Features
- 2D Electrophoresis Simulation complete with protein-specific database searches
- Integrated and modular Mass Spectrometer simulation.