Link Windows 32
Link Windows 64
Link Mac
Link Android
Link IPhone
MedInria is a multi-platform medical image processing and visualization software. It is free and open-source. Through an intuitive user interface, medInria offers from standard to cutting-edge processing functionalities for your medical images such as 2D/3D/4D image visualization, image registration, diffusion MR processing and tractography.
Detailed Features of medInria:
MedInria can be used for a wide variety of tasks. It provides in its current release 6 main feature categories including:
- Database management and file import: DICOM read, PACS interrogation, various file formats handling, etc.
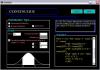
MedInria comes with a patient centric database, accessible in the Browser. It provides the usual features related to data management: import or index data from various data sources (file system, Dicom, external databases - Shanoir, PACS server), export data to disk. It is able to load any format that is supported by ITK and GIS images. Finally, it also allows for a quick preview of data in your database through thumbnails.
- Visualization: 2D to 4D image visualization, tensor, surfaces visualization in a drag-drop and tabs interface
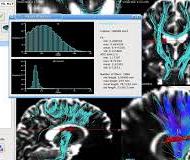
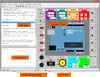
MedInria provides a powerful environment to visualize many kinds of medical data. You may view typical 2D/3D images, side by side or superimposed on separate layers. We also provide the usual slice by slice navigation as well as 3D/4D volume rendering of images. All VTK meshes and line sets are also supported and may be viewed on top of other images, providing a comprehensive way e.g. to display cardiac models over the corresponding CT or brain surfaces on top of their native MR image. Among other possibilities, 4D data is also supported, both for meshes and images, providing a great way to observe at the same time simulated models and real images to see how they fit together. Finally, medInria provides tensor and fibers visualization for diffusion imaging.
- Diffusion images processing: from tensor estimation to tractography, including the visualization of tensor fields and bundling of fiber tracts
Diffusion imaging and tractography are of great interest to better understand the brain organization and its alteration by diseases. Diffusion imaging processing in medInria allows to go from the raw diffsuion data acquisition to the extraction of fiber bundles : computation and visualization of tensor volumes, extraction of scalar indicators such as FA or MD, whole-brain tractography, filtering and tagging of fiber bundles, etc.
- Image segmentation: manual interactive (and soon automatic) segmentation of images
MedInria provides a new interface to perform image segmentation. As of now, it includes a paintbrush tool to interactivaly paint a segmentation on the image and export or save it ot the database. It will include many more automatic algorithms in the future for various applications.
- Registration of images: register images utilizing either linear or non-linear transformations
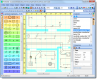
We provide an entire interface to virtually any registration method. As of now, registration in medInria incorporates efficient methods for the linear and non linear registration of anatomical images. Among them are block-matching linear registration, Optimus (Newuoa for MI-based rigid registration), and diffeomorphic demons non linear registration. All these algorithms are included in an interface for side by side comparison of images, or fused comparison to ealuate the registration quality.
- Filtering of images: Apply basic or advanced filters to an image (from some ITK-based basic filters to more advanced denoising algorithms)
Last but not least, medInria aims at providing tools (basic and cutting-edge) for preprocessing and enhancing medical images. This feature is provided by the filtering workspace, which includes a side by side comparison interface with the usual controls. The filters in medInria include basic filters such as median filter or Gaussian blur, as well as more advanced ones such as the NL-Means denoising method.